Let's come to the so-called "Bacteria"....
The existence of bacteria is indisputably true, so postulated by some.... well, maybe they exist, maybe they don't... the fact is that no bacterium has ever actually been isolated, so that you could say you can physically grasping or working with it - what can be established with 100% certainty is the fact that there are NO pathogenic so-called “bacteria”, because THAT is the same nonsense as with the imaginary, non-existent “viruses”!!!! And so-called “studies” WITHOUT specifying the method (the so-called “isolation”) are to be classified as fraud from the outset anyway!!!
https://www.scielo.br/j/rimtsp/a/JvmbVCwyS65VHxLr7dpqjYb/?lang=en
Whereby the first sentence in the “Abstract” already says all the nonsense!
The word “subculture” means that the so-called “strains” used come from a previous fraudulent culture!!!
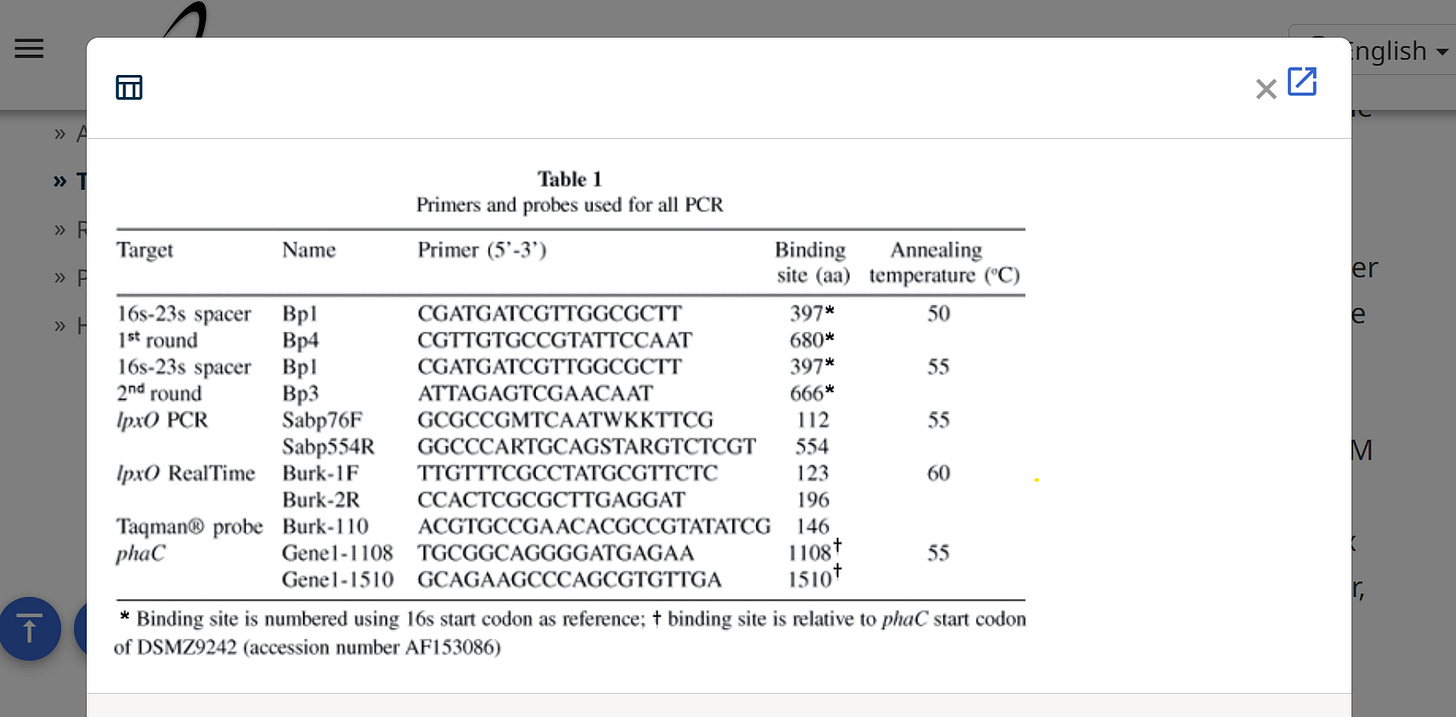
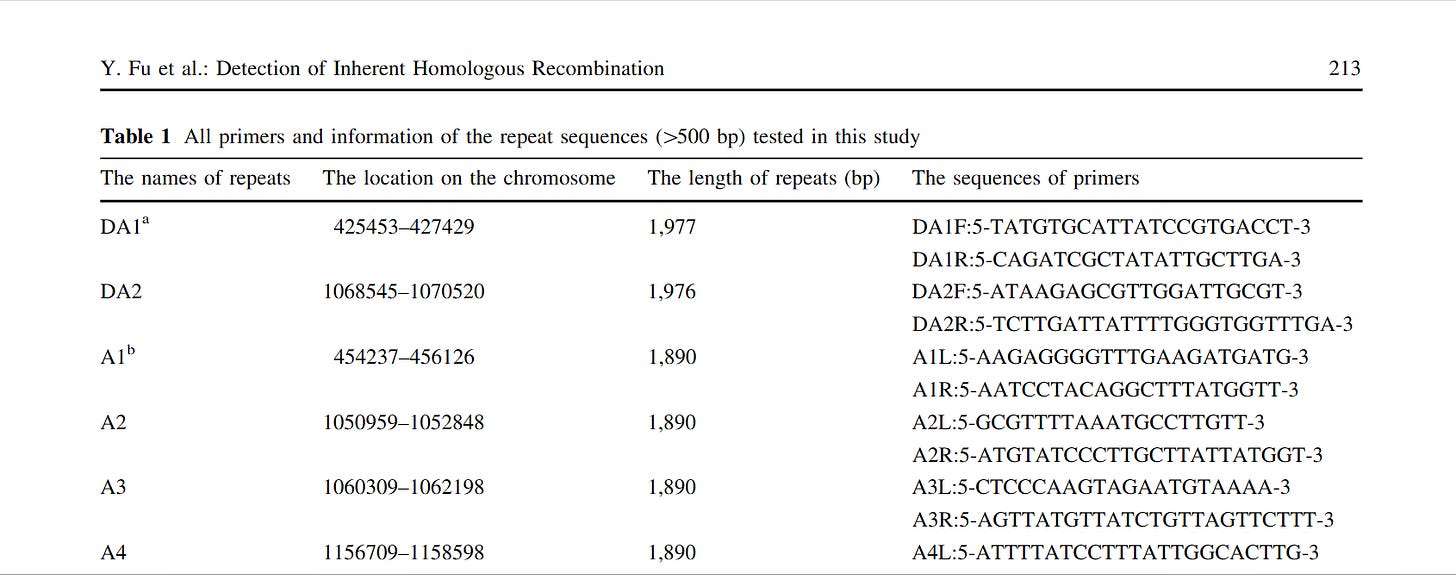
PCR targets, mixes and cycling conditions:
All primer design was conducted using Primer Express software (Applied Biosystems). Table 1 shows the primer pairs used in this study, their binding site relative to the start codon of the gene of interest and the annealing temperature for their respective PCR programs. The Taqman probe used for realtime detection lpxo was synthesized by Biosearch Technologies Inc. (Novato, Ca 94949, USA) and consisted of a 5' 6-FAM reporter label and a 3' 'Black Hole Quencher'. Semi-nested PCR was performed as described previously7. phac PCR was performed using the following PCR mix conditions; 1 unit(u) Amplitaq Gold Enzyme (Applied Biosystems), PE PCR buffer, 1.5 mM MgCl20.25 MM each dNTP and 0.5 0.5 μM of each primer in a total volume of 20 μL (including 8 8 μL of template). lpxo PCR was performed using the following PCR mix conditions; 0.5 u Amplitaq Gold Enzyme (Applied Biosystems), PE PCR buffer, 2.5 mM MgCl20.2 MM each dNTP and 0.4 0.4 μM of each primer in a total volume of 20 (including 8 8 %L of template). Realtime lpxo PCR was performed using the following PCR mix conditions; 0.75 u Amplitaq Gold Enzyme (Applied Biosystems), PE PCR buffer, 4.0 mM MgCl2, 0.2 mM each dNTP and 0.2 0.1 0.1 0.2 1 1 1 1 20 (including 8 8 LL of template). All other conventional PCR methods were performed under the following conditions, pre-PCR of 10 minutes at 95oC to fully denature the template DNA and activate the polymerase followed by 45 cycles of 30 sec at 94oC for denaturation, 30 sec at the appropriate annealing temperature and 45 sec at 72oC for extension. Following the final cycle the samples were maintained at 72 C for a further seven min. PCR products were demonstrated by Ethidium bromide gel electrophoresis on 2.5% agar gels. Digital gel images were captured and optimized for brightness and intensity using UVIdoc capture system (Cambridge, UK). The lpxorealtime PCR protocol was performed under the following conditions, pre-PCR of 10 minutes at 95oC to fully denature the template DNA and activate the polymerase followed by 50 cycles of 10 sec at 94oC for denaturation, 10 sec at the appropriate annealing temperature and 30 sec at 72oC for extension. Amplification and detection was carried out in an Applied Biosystems Prism 7000 Sequence Detection System.
https://semmelweispharma.com/netcorefacility/images/ABI_Prism_7000_Sequence_Detection_System_User_Guide__1_.pdf
And now we can choose where these so-called “dead cell debris” belong...
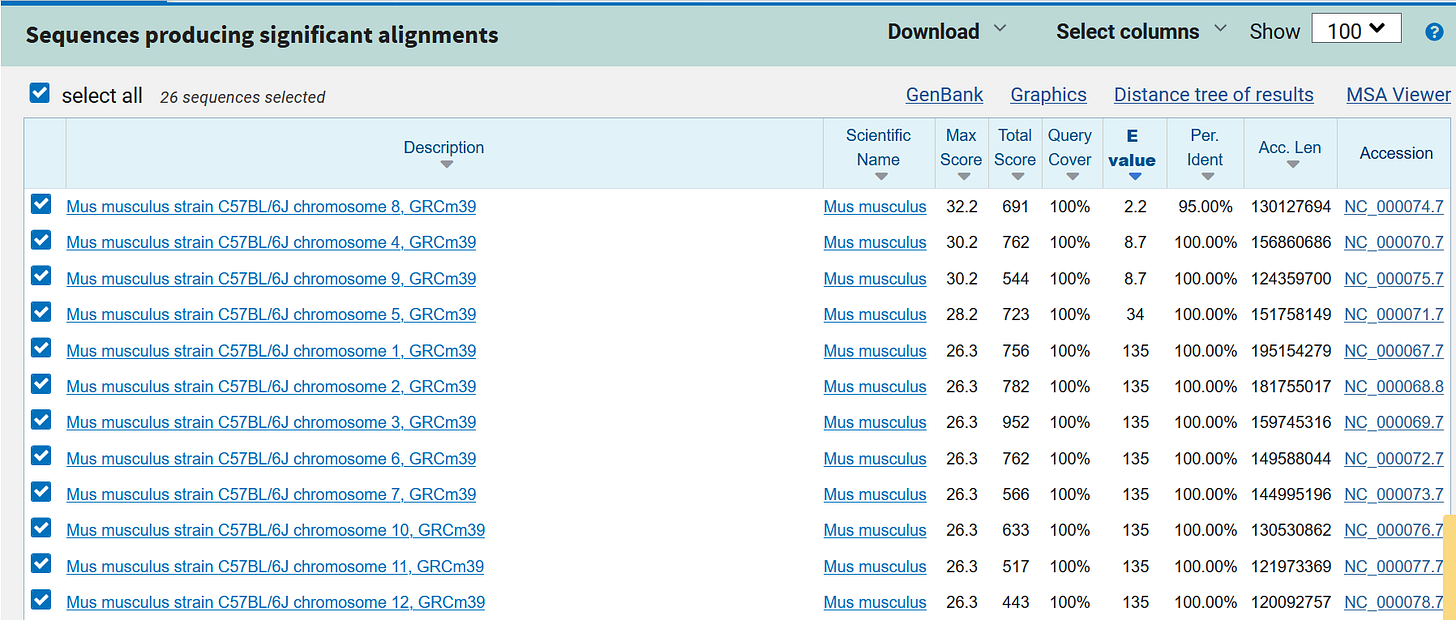
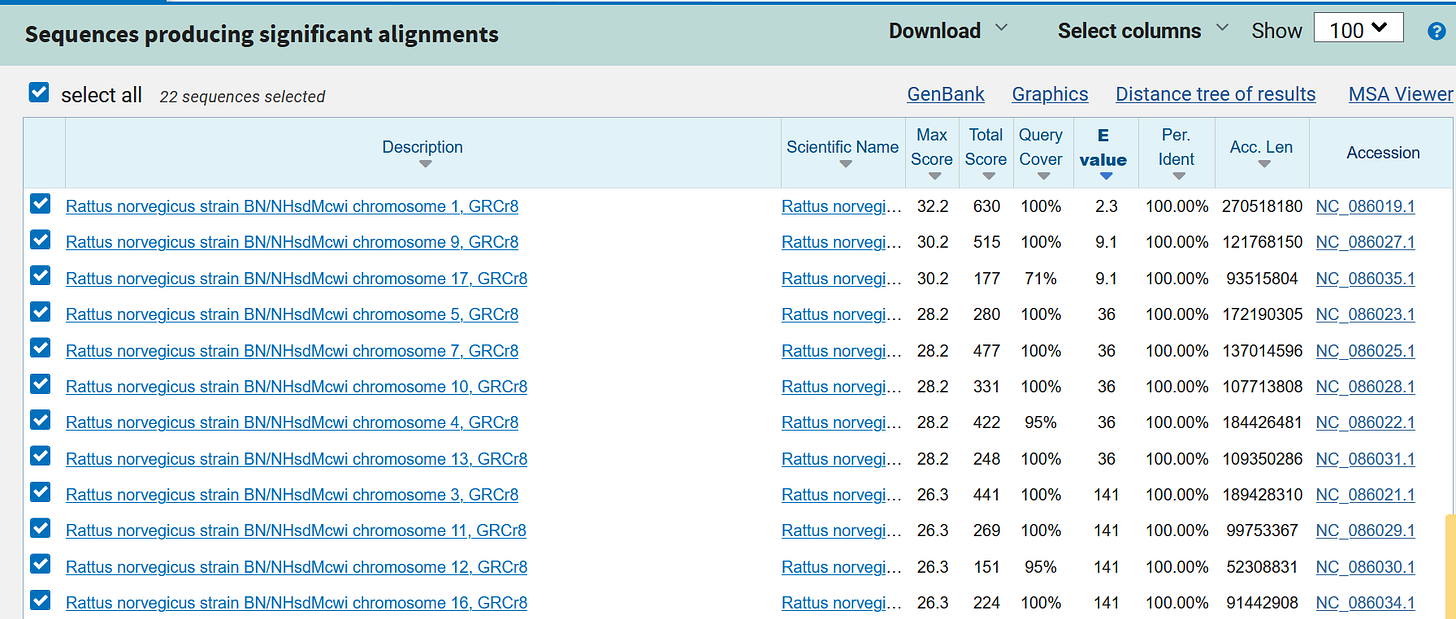
e.g. the “letter group” TTG TTT CGC CTA TGC GTT CTC
https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=OGP__9606__9558&LINK_LOC=blasthome
https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=OGP__10090__9559&LINK_LOC=blasthome
https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=OGP__10116__10621&LINK_LOC=blasthome
And when it comes to the so-called non-existent “microbes”, the list is endless, so you can choose whatever you need....
https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=MicrobialGenomes
etc. etc.
and of course with magic aids such e.g. as that:
https://thalljiscience.github.io/BioDoc.pdf
or this - The primer software from “Applied Biosystem”:
https://www.bu.edu/picf/files/2010/11/Primer-express-30.pdf
or here:
https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_041428.pdf
or this:
https://microbeonline.com/api-for-microbial-identification/
etc.
And then of course we can still read all sorts of other nonsense… blah blah blah!
A screenshot taken a long time ago
- and if you consider all the chemical substances that have been used to manipulate the weather for decades, then no one should be surprised at what comes down with the rain etc. and how the soils are already damaged by pesticides.....
And this whole hocus pocus was also the same in the past!
https://journals.asm.org/doi/pdf/10.1128/jcm.33.8.2131-2135.1995
And now a little more about so-called “antibody production”, which makes your stomach churn when you read it and think about the fact, that people and animals are administered this....
https://apjai-journal.org/wp-content/uploads/2018/02/8ProductionofSpecificMonoclonalAntibodiesAPJAIVol14No1June1996P43.pdf
on page 2 at the bottom you can read the “Freund's adjuvant” - and here is the safety data sheet:
https://assets.thermofisher.com/TFS-Assets/LSG/SDS/RF0816_MTR-NARF_EN.pdf
And here's a little more of the so-called “bacteria”(with the usual blah blah blah) that don't exist, THAT'S A FACT!!!
https://www.researchgate.net/publication/256540850_Detection_of_Onchocerca_volvulus_in_Latin_American_black_flies_for_pool_screening_PCR_using_high-throughput_automated_DNA_isolation_for_transmission_surveillance
etc.
here the “letter groups” are really flying around your ears…😂
https://pmc.ncbi.nlm.nih.gov/articles/instance/334508/pdf/nar00234-0094.pdf
https://softgenetics.com/products/genemarker/t-rflp/
https://softgenetics.com/PDF/T-RFLPapplicationnote.pdf
and then of course you need a tool to analyze the RFLP data....😉
https://www.rdocumentation.org/packages/RFLPtools/versions/2.0
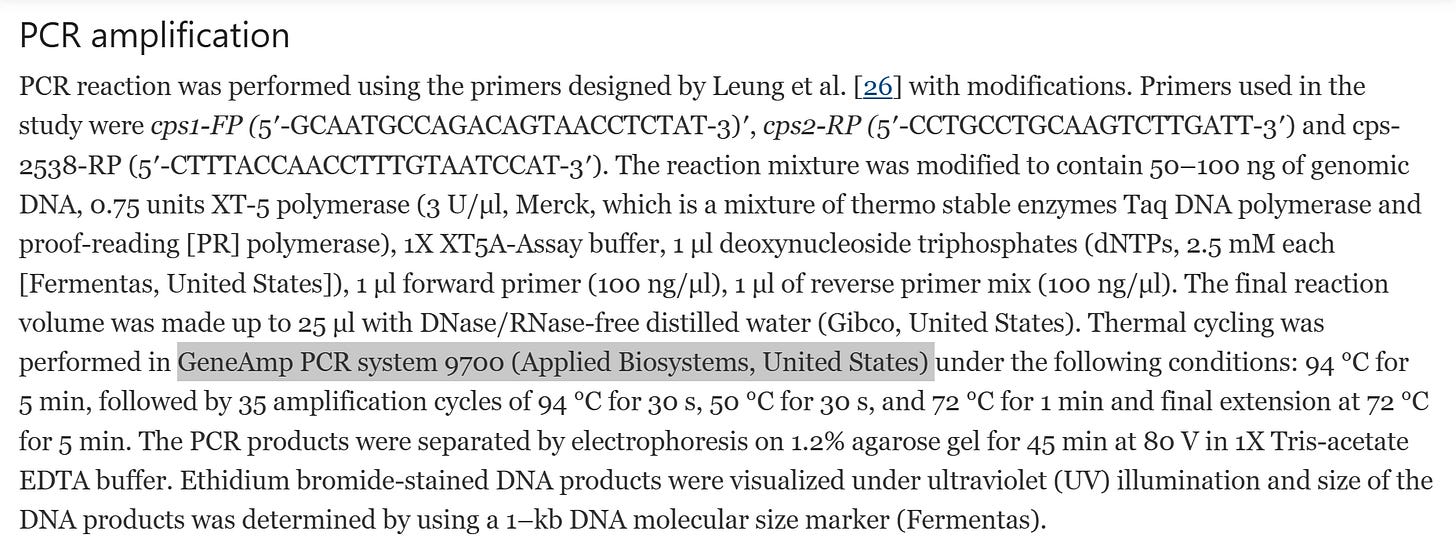
https://pneumonia.biomedcentral.com/articles/10.1186/s41479-017-0032-3
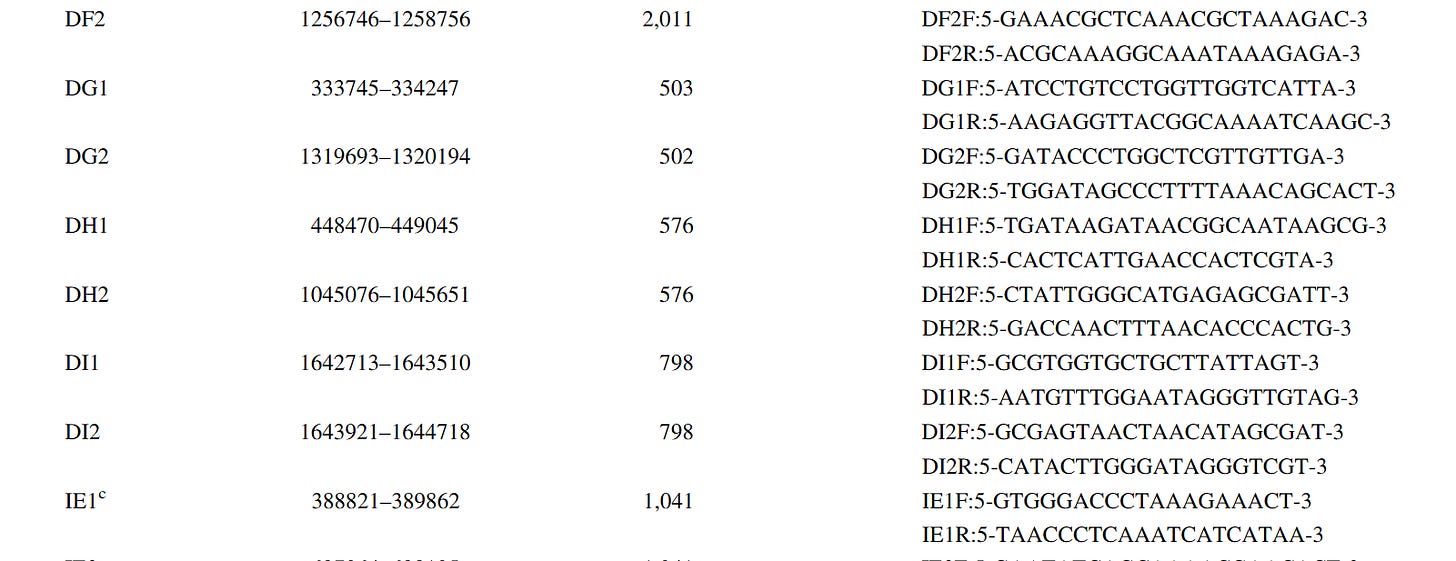
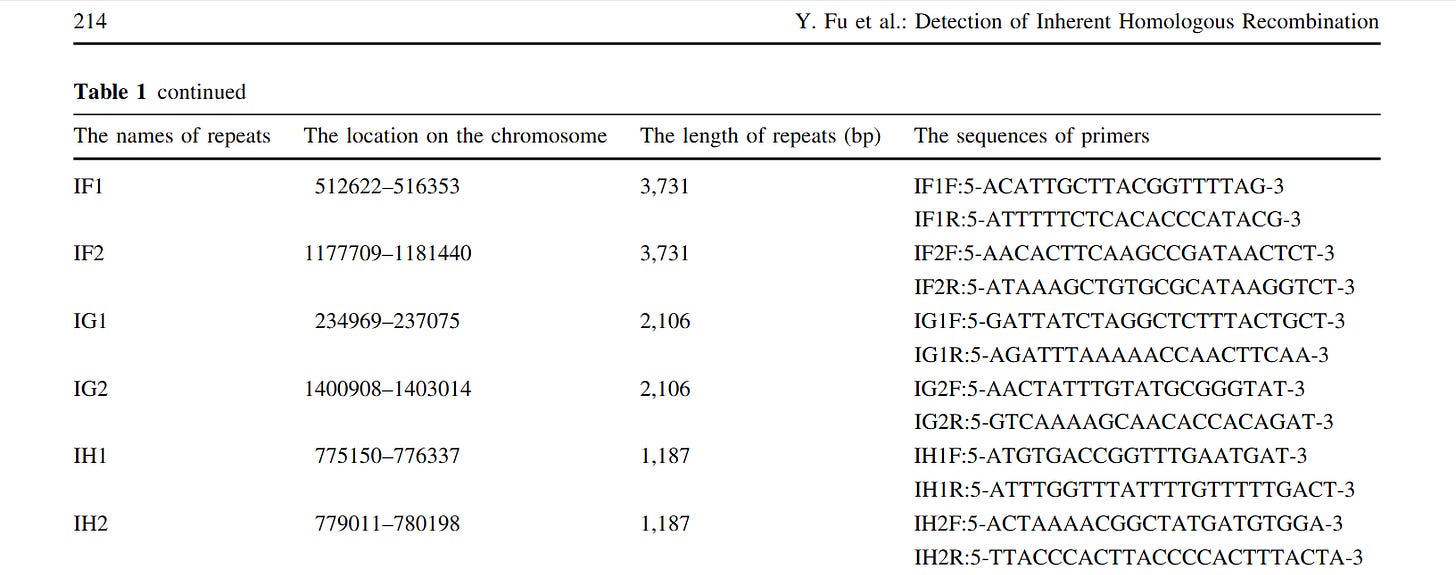
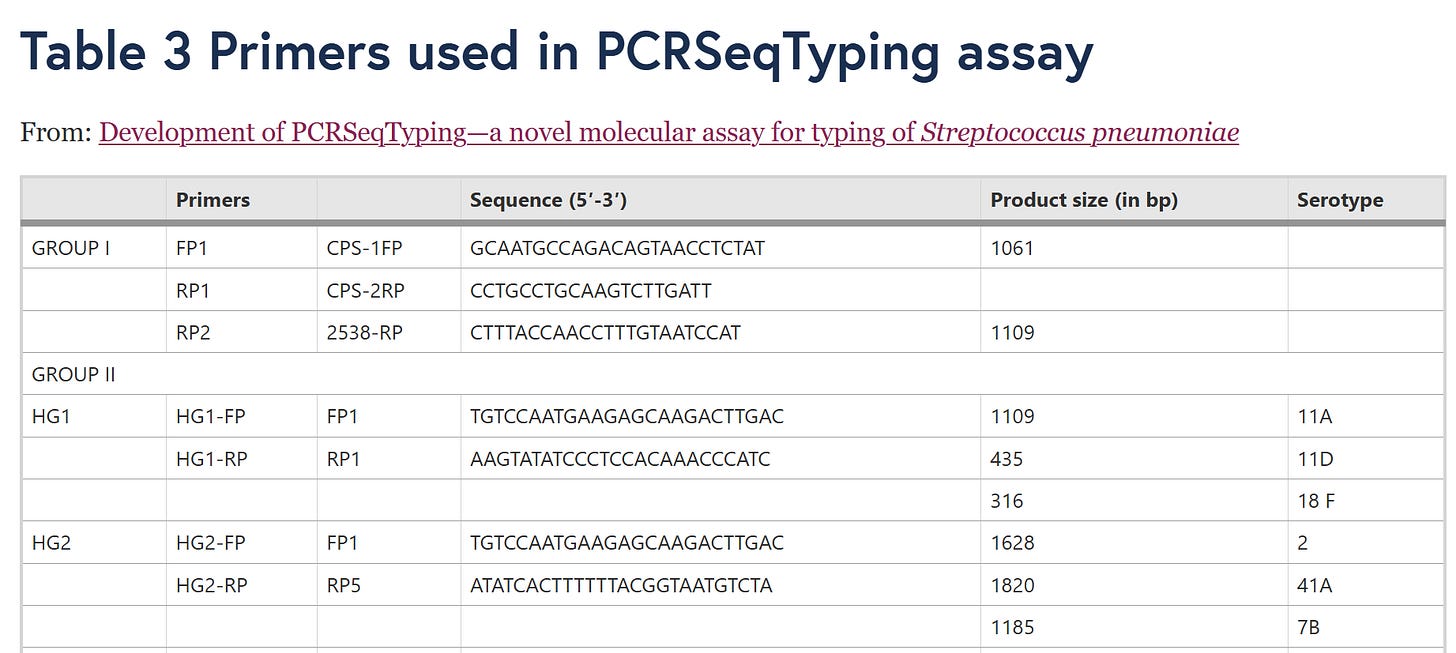
In Table 1, “PCRseqtyping results for 91 SSI strains” means that the same nonsense has already been done there by PCR sequencing, so this is also a “subculture”!
https://www.labx.com/product/abi-geneamp-9700
https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_040970.pdf
https://cores.research.asu.edu/sites/default/files/2019-04/ABI%203730XL%20manual.pdf